Infrastructure to support human genomics data management
Managing human genome data using Gen3
What is Gen3?
What groups in Australia are using Gen3?
How do I establish/deploy a Gen3 instance?
How do I develop a Gen3 data dictionary?
Creating Virtual Cohorts across multiple genomic repositories Background - Virtual Cohorts
National community needs - Virtual Cohort Assembly (2022)
The GA4GH Beacon protocol which enabled discovery of virtual cohorts across multiple sites
GA4GH Beacon implementation guides
[section 3] Helping automate the work of Data Access Committees (DACs) Background - DAC automation
National community needs - DAC Automation (2022)
REMS and its deployment in AWS
Experiences using the REMS (Resource Entitlement Management System) software package
[section 4] Federated Identity & Access Management (IAM) Background - Federated Identity and Access
National community needs - Federated IAM (2022)
CILogon and COmanage for federated access management in Australia
CILogon and how it enables identity management
[section 5] Human Genomic Data and Metadata Archiving Background - Human Genome Data Archiving
Managing human genome data using Gen3
What is Gen3?
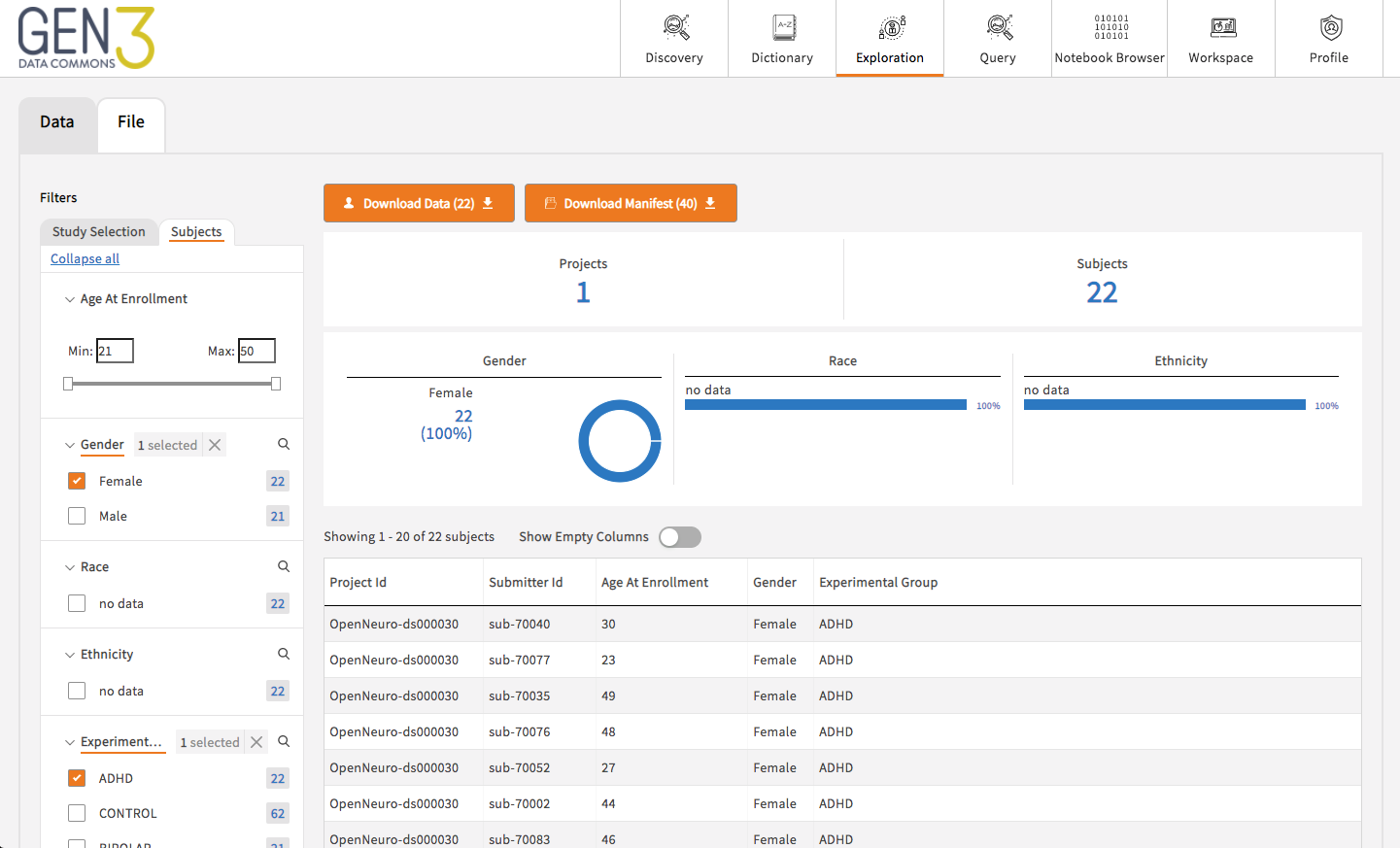
Gen3 (https://gen3.org/) is an open source software suite that allows data to be received, managed, described, quality controlled and shared with authorised individuals.
It's tech that is used to underpin numerous very large US NIH-funded human genomics efforts that collectively house and describe data from 100s of 1000s of samples:
Powered by Gen3
It provides a framework to organise files, and responsive interfaces to allow searching of data collection contents:
Storage of the data files can be held across various object stores.
Access to the actual genomic data is managed by a Data Access Committee for each datasets, and controlled by various tools whih are part of the Gen3 system.
A Gen3-based storage system can be set-up by the administrators to allow direct data downalod, or it can be set up to enable linking to cloud-based analysis systems.