News
Subscribe to the Australian BioCommons monthly newsletter or read previous editions
Thousands join international Galaxy training events
The Galaxy platform is celebrating its 20th anniversary this year and Galaxy Australia is one of the core BioCommons services. We are proud to provide and support a variety of opportunities for researchers to engage with the team, community and tools.
The Galaxy platform is celebrating its 20th anniversary this year. This collaborative data analysis platform is widely used by scientists around the world, and underpins many computational biology services. Galaxy Australia is one of the core services BioCommons delivers, so naturally we provide lots of opportunities for researchers to engage with the team, community and tools.
The Galaxy Australia team recently supported the Singapore Biology League, who chose to incorporate Galaxy into their online collaborative biology contest for the first time. This massive event welcomed over 2000 pre-university students who formed teams to run 2054 tools on Galaxy Australia over 4 hours. It was a great opportunity to broaden participants’ exposure to biology and bioinformatics beyond the school curriculum.
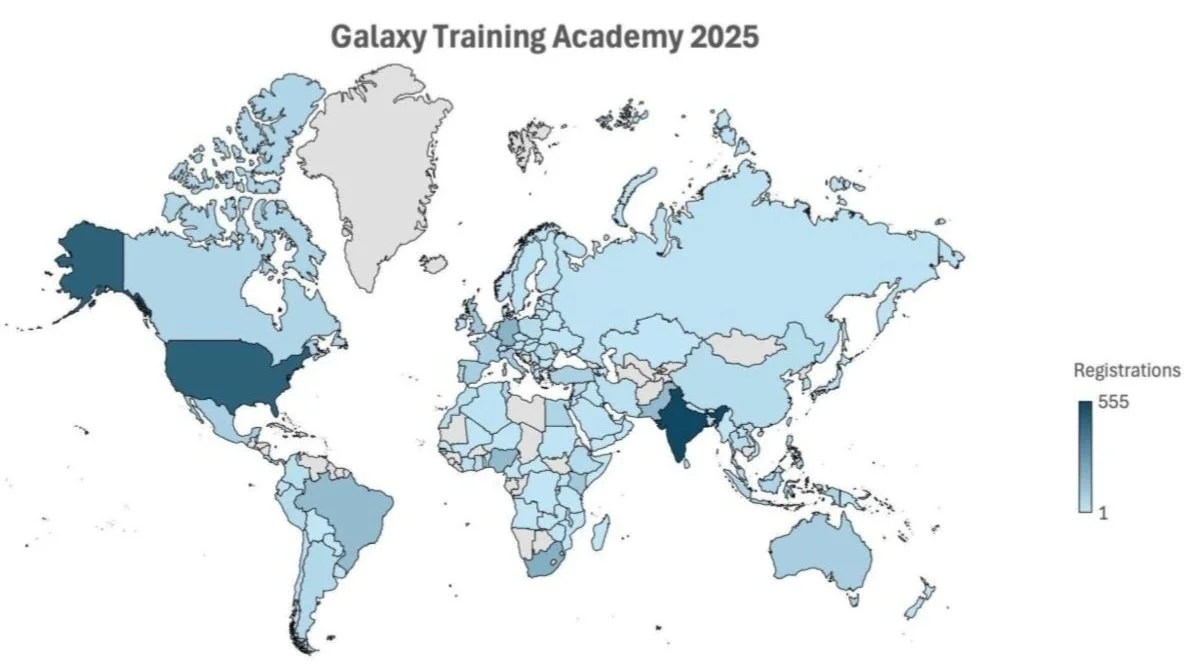
The week-long Galaxy Training Academy attracted more than 3500 people from across the globe this year. Seventy three Australian researchers were amongst the international cohort joining from their homes and offices to work through tutorials, with support as they needed it. Live help was provided in our time zones by Galaxy Australia’s Dr Anna Syme, Dr Igor Makunin and Dr Tristan Reynolds, who fielded questions on a wide range of topics from fungal genomics to bat ecology! Participants brought their own specialities as they learned how to use the fully subsidised Galaxy Australia platform for proteomics, genome assembly, transcriptomics, single cell RNAseq, microbiome analysis, machine learning and more.
In the lead up to the Galaxy Training Academy, BioCommons’ Dr Tiffanie Nelson and Galaxy Australia’s Dr Tristan Reynolds helped researchers understand what’s possible by presenting the webinar: No code, no problem: data analysis for biologists with Galaxy Australia. Examples included how Galaxy Australia is being used for biosecurity screening, foodborne pathogen detection and building reference genomes of the critically endangered swift parrot, as well as a tour of the practical features of Galaxy Australia that make sophisticated workflows like these accessible to all, regardless of their computational skills.
Get started by exploring the tutorials available: Galaxy Training Network
If you’re an existing Galaxy Australia user, improve your methods with our Top Tips videos
Stop the rot! Plant Bacteriologist’s genome assembly becomes Galaxy Australia’s 11 millionth job
An enthusiastic new user recently submitted the lucky 11 millionth data analysis job to the Galaxy Australia platform. Plant Bacteriologist Dr Toni Chapman has begun regularly using the fully-subsidised service for her genome assemblies of bacteria important to agricultural plant biosecurity and production.
Image courtesy of Dr Toni Chapman
An enthusiastic new user recently submitted the lucky 11 millionth data analysis job to the Galaxy Australia platform. Plant Bacteriologist Dr Toni Chapman has begun regularly using the service for her genome assemblies of bacteria important to agricultural plant biosecurity and production.
As a Senior Research Scientist in Agriculture and Biosecurity at the NSW Department of Primary Industries and Regional Development (DPIRD), Toni’s work spans both diagnostics and research. Using NovaSeq and HiFi sequencing technologies, she assembles bacterial genomes to help growers diagnose the cause of disease symptoms in their plants or to identify bacteria present in suspect samples collected by biosecurity officers.
In the past, Toni needed to rely heavily on bioinformaticians for assistance with genome assembly of the bacteria she works with. Now after attending a training workshop and signing up for access to the fully-subsidised Galaxy Australia service, she independently completes more of the analysis steps and is able to identify the bacterial pathogens herself.
“Over time I have been learning to code and to use various programs for genome assembly, but only returning to it on a casual basis makes it very hard to maintain the necessary skills in the bioinformatics space. Being able to design workflows in Galaxy Australia that I can come back to at any time makes assembling genomes easy, even if there has been weeks or months between visits.”
Toni contributes to the Plant Pathogen ‘Omics Initiative, which is generating molecular reference data for plant pathogens in Australia. Established by Bioplatforms Australia to support research in plant protection and enhance biosecurity surveillance efforts, the national plant pathogen community is collaborating to create high quality data that can be shared with all researchers via the Data Portal. Toni was one of a large group that came together with members of the Functional Fungi and Plant Pathogen ‘Omics National Initiatives for a hands-on bioinformatics workshop, learning how to use Galaxy and the programs needed for genome assembly using their own real-world scenarios.
As part of her contribution to the Initiative, Toni is conducting genome assembly on Pseudomonas species that cause disease in plants, to update the taxonomy of collection isolates and gain a more accurate view of which pathogens are in Australia. In another project, she is sequencing the bacterial pathogens that cause soft rot disease in plants. The incidence of soft rot is increasing, and so is the range of bacteria that can cause the disease. The newly assembled genomes of these bacteria are updating existing culture collection identities and helping to understand the diversity of bacteria that cause soft rot infections in Australia.
If you’d like to find out more about her work, see Dr Toni Chapman’s publications.
If you are interested in hearing about the different types of research that Galaxy Australia gets used for, watch this recorded webinar: No code, no problem - data analysis for biologists with Galaxy Australia.
An Australian community for computational structural biology
A passionate group of structural biologists has formed the Australian Structural Biology Computing Community, to share computational knowledge, methods, and resources.
The active new community is receiving support from a range of partners and advocates, including L-R Johan Gustafsson (BioCommons), Steven Manos (BioCommons), Kate Michie (UNSW) and Andrew Gilbert (Bioplatforms Australia)
The explosion of possibilities presented by deep learning approaches in structural biology research has created many new opportunities and challenges. A passionate group of structural biologists has formed the Australian Structural Biology Computing Community, to approach this new era as part of a community that shares computational knowledge, methods, and resources.
This community-driven approach brings together a diverse group of people, with initial contributions forming around leads from the Structural Biology Facility at UNSW, and an academic panel of experts from Monash University, Walter and Eliza Hall Institute of Medical Research (WEHI), University of Western Australia (UWA), Australian National University (ANU), Bio21 Institute of Molecular Science and Biotechnology (Bio21), University of Melbourne, La Trobe University, University of Queensland (UQ) - IMB, University of Sydney, Griffith University, Swinburne University of Technology, CSIRO, and the University of Adelaide. Anyone involved in structural biology in Australia is invited to join and there are lots of different ways to get involved.
The Community for Structural Biology Computing in Australia webpage is a useful new resource for all users of computing for structural biology research in Australia. The page is constantly evolving and expanding, and it currently focuses on the use of deep learning methods in Structural Biology. It includes practical guides on topics like “Best practices for presenting and sharing AlphaFold models in a paper” as well as news items and announcements for relevant courses and meetings.
Australian BioCommons supports the community by hosting quarterly online meetings that aim to tease out how computational structural biologists’ challenges might be addressed with community-scale responses and national research infrastructure solutions. If you join the mailing list via the community webpage, you will receive updates and invitations to community meetings and the discussions in Slack.
BioCommons began providing broad, fully subsidised, access to structural prediction in 2022 by making AlphaFold2 available within its Galaxy Australia service. The Australian AlphaFold2 Service provides both an easy-to-use interface and dedicated GPUs to Australian researchers. When BioCommons hosted the international 2023 Galaxy Community Conference, the keynote speech by Chief Scientist of the Structural Biology Factility at UNSW, Kate Michie, generated much excitement around forming an Australian community of practice for computational structural biology as an avenue for collectively addressing the challenges presented by deep learning in structural biology.
BioCommons has supported key research stakeholders to refine the new community’s purpose, began running quarterly community meetings, and helped to establish the shared community spaces like the Australian Structural Biology Computing website and GitHub. As well as facilitating consultations with infrastructure partners and the broader computational infrastructure community, a group of national panel of experts has been identified.
This community collaborates with their peers to:
Collectively create and maintain community forums and centralised collaboration platforms to support collaboration and knowledge sharing (i.e. methods and documentation);
Foster collaboration between structural biologists, computer scientists, and data scientists, thereby creating interdisciplinary teams to help tackle complex challenges, validate results and ensure robust applications of deep learning methods;
Lead the review, prioritisation, testing, optimisation, and sharing of deep learning codes, software and approaches that are of broad relevance and interest to the Australian research community;
Develop quality assessment tools to help evaluate the quality of calculated structures, and help guide researchers towards reliable predictions; and,
Address the ethical implications of AI-driven structural predictions, as well as discuss transparency, bias and interpretability to ensure responsible use of these technologies.
The Australian Structural Biology Community is poised to tackle a set of pilot activities aimed at fast tracking a national response to the challenges facing computational approaches in structural biology. A much anticipated future output is an infrastructure roadmap document that will formalise and describe the high level requirements of the community. This collaborative effort between the new Australian Structural Biology Community, Australian BioCommons, and BioCommons infrastructure partners will support the Australian Structural Biology community as new needs arise relating to bioinformatics tools, software, infrastructure or training.
Keep in touch by subscribing for updates at the Community for Structural Biology Computing in Australia webpage.
Repurposed hardware boosts national capacity and powers innovation
QCIF Ltd has made high-performance hardware available to the Australian BioCommons, giving the hardware a second life and uplifting national capacity for running AlphaFold 2 jobs in Galaxy Australia while supporting innovation through other GPU-enabled tools.
This story is co-published with QCIF Ltd
After successfully completing a previous project, QCIF Ltd made available high-performance hardware to the Australian BioCommons, giving the hardware a second life in enabling research and uplifting national capacity for the benefit of the scientific community.
Well suited for running AlphaFold 2 jobs, the five General-Purpose Graphics Processing Units (GPGPUs) are now being used to enhance the national compute network behind the Galaxy Australia service.
The impact of this repurposing goes beyond infrastructure improvements. It has significantly expanded Galaxy Australia's capacity to support research and innovation by enabling the use of other GPU-enabled tools that offer major benefits to the scientific community. GPU processing can provide massive improvements in computational efficiency, decreasing processing times to less than 5% of conventional equivalents.
Dr Cameron Hyde, a bioinformatician at QCIF who supports the development of national software platforms like Australian BioCommons' Galaxy and Apollo services, co-authored the original AlphaFold 2 wrapper that enabled the tool to run within Galaxy Australia, ensuring both a friendly user-interface as well as instant access to the GPU clusters required to power the tool. He shared his enthusiasm for the new possibilities unlocked by the repurposed hardware which was originally part of an investment made in 2021 by the Australian Research Data Commons (ARDC) to support national platform projects and now directly enhances the bioinformatics services he helps deliver to Australian researchers. “Now that we have five GPU nodes of our own, we have room to experiment and explore new GPU-enabled tools. This gives us room to innovate beyond AlphaFold and accelerate scientific discovery in other research domains.”
For example, Galaxy Australia’s lead Bioinformatician Michael Thang has been using the hardware to explore running Nanopore’s “Dorado” on Galaxy Australia. Dorado is a high-performance basecaller for Oxford Nanopore Technology sequencing data. This innovation would enable researchers to conduct their entire analysis, from raw sequencing data through to assembled genome, all within the Galaxy Australia service.
Collaboration driving innovation
Developed by Google DeepMind, AlphaFold is an AI system that predicts a protein’s 3D structure from its amino acid sequence with accuracy comparable to experimental methods. In 2020, Australian BioCommons identified an opportunity to democratise access to this powerful tool by making AlphaFold 2 available through Galaxy Australia. This gave Australian researchers much greater accessibility to AlphaFold 2, allowing life scientists to easily visualise proteins in a manner inaccessible to all but dedicated structural biology researchers. This advance has supported research into protein-protein interactions, activation and inhibition mechanisms, and drug design.
By 2025, use of AlphaFold 2 has surged, evolving from an analytical tool for individual proteins into a routine screening tool for studying protein-protein interactions. To support this shift, Dr Hyde collaborated closely with Australian Structural Biology Computing Community to develop extensions to the AlphaFold Galaxy tool, including new output formats, input parameters, and an option to re-use intermediate files for improved efficiency.
Supported by the Australian BioCommons, AARNnet, QCIF Ltd, and The University of Melbourne, the optimised system now provides fully subsidised access for all Australian researchers via the Australian Alphafold Service. We extend our sincere thanks to the Australian Research Data Commons (ARDC) for providing the hardware to QCIF Ltd and enabling its reuse by Australian BioCommons.
Microbiology Lab empowers microbial data analysis with integrated tools, workflows and compute
Microbiology Lab offers a customised, user-friendly view of Galaxy Australia that provides rapid access to popular tools, workflows and compute for analysis and visualisation of microbiomes and omics data from microbial isolates.
Galaxy Australia’s Microbiology Lab is now providing researchers with rapid access to popular tools, workflows and compute for analysis and visualisation of microbiomes and omics data from microbial isolates.
Development of this curated view of Galaxy Australia was driven by the international microbial research community who identified the most commonly used tools in the field through meticulous research, surveys and community consultations.
The Lab integrates more than 220 tools and 65 workflows with step-by-step tutorials and structured learning paths for a suite of different analyses. Microbiology Lab pairs perfectly with the computing power of Galaxy Australia, which is underpinned by computational resources provided by AARNet, ARDC Nectar Research Cloud, the University of Melbourne, QCIF, Pawsey Supercomputing Research Centre, National Computational Infrastructure, and Microsoft Azure. From raw data, to differential analysis, visualisation and assembly, Microbiology Lab makes it easy to get started with reproducible analysis of microbial data.
Galaxy Australia’s Microbiology Lab is the latest release in a series of Labs supporting research domains. It represents an important step forward in Australian BioCommons activities to support Australian microbial research. The Microbiome Analysis Infrastructure Roadmap for Australia identified a need to implement a shared platform that eases access to preferred tools and workflows for analysis of microbial data with sufficient computational power. This fully-subsidised resource is expected to improve efficiency for researchers.
If you are an Australian researcher with an interest in microbial isolates and microbiomes, be sure to take a tour of the Galaxy Australia Microbiology Lab and try it out now!
Join 41,000 users of this fully-subsidised and easy to use Australian bioinformatics platform
Galaxy has proven to be such a versatile data analysis platform that Galaxy Australia supports over 41,000 users. Now is the perfect time to get on board with Galaxy, with a variety of ways to get involved on offer in the coming months.
Galaxy has proven to be such a versatile data analysis platform that Galaxy Australia now supports over 41,000 users. This fully subsidised, open-source system for analysing and visualising data, authoring workflows, training, publishing tools, and managing infrastructure is powered by a world-wide community of 500,000 Galaxy users as well as the institutions who invest in its growth. Now is the perfect time to get on board with Galaxy, with a variety of ways to get involved on offer in the coming months.
If you are curious to see how Galaxy is being used in a wide range of research settings, come along to our upcoming webinar showcasing how Galaxy Australia is supporting genome assembly and annotation, metagenomics, proteomics, transcriptomics, data visualisation and more. Register now for No code, no problem - data analysis for biologists with Galaxy Australia on Tue 29 Apr 2025.
The global community’s annual training event - Galaxy Training Academy - is coming up in May. This week-long, completely free, online training event will help you master the Galaxy platform for data analysis. It is self-paced and you can choose from topics including Proteomics, Assembly, Transcriptomics, Single Cell, Microbiome or Machine Learning data analysis in Galaxy. The Galaxy Australia team will be on hand to answer any questions you have during the event over 12-16 May.
Most people simply make use of the platform to do their research, but there’s an open invitation to contribute to this lively community too. The international Galaxy Community,has put together a Get Started page to help you find out more about terminology, resources, servers, types of communities, and the different ways to contribute to the great things the Galaxy community achieves together. You can also connect with the community in person at the Galaxy and Bioconductor Community Conference 2025 is coming up in June in the USA.
Strategic partnerships formed around the platform are key in making it readily available to Australian researchers. Australian BioCommons is able to offer the service at no cost to researchers because of the commitment of valuable partners QCIF, The University of Melbourne and AARNet who help manage the Galaxy Australia service and provision of computational resources provided by respected national institutions and providers AARNet, ARDC Nectar Research Cloud, the University of Melbourne, QCIF, National Computational Infrastructure, Pawsey Supercomputing Research Centre and Microsoft Azure. These efforts are backed by funding from the University of Melbourne, the Queensland state government and Bioplatforms Australia, who bring Federal investment through the National Collaborative Research Infrastructure Strategy.
If you’re ready to start using Galaxy Australia, you can jump right in and try it now: usegalaxy.org.au/
Galaxy Australia’s 10 millionth job relied on popular locally-developed tool
The ten millionth job completed on the Galaxy Australia service analysed pathogen samples for known antibiotic resistance genes using the ABRicate tool. You’d be surprised how many time this Australian tool gets used.
At any one time, there is a wide array of research being done using Galaxy Australia. When the ten millionth job was submitted to the service recently, it illuminated the importance of good bioinformatics software. The particular job was one of a bunch that analysed various pathogen samples for known antibiotic resistance genes.
Over the last 7 years, more than 40,000 users representing researchers, students, trainers and trainees have demonstrated the platform's versatility across a wide range of fields. This milestone job investigating antimicrobial resistance employed the ABRicate tool, authored by Australian bioinformatician Torsten Seemann. Further investigation showed that 490,507 jobs run on Galaxy Australia also used ABRicate in their workflows!
The University of Melbourne’s A/Prof Torsten Seemann is the Lead Bioinformatician at the Centre for Pathogen Genomics and the Director of Bioinformatics at Doherty Applied Microbial Genomics. As a world-renowned bioinformatician he has developed cutting edge analysis approaches to enhance the use of genomic data. Torsten has authored many popular software tools for the analysis of bacterial sequence data, particularly in the fields of public health and clinical microbiology. He is also a strong advocate for open-source software and open science.
He created ABRicate for the mass screening of assemblies for antimicrobial resistance and virulence genes. Given a FASTA contig file or a genbank file, ABRicate will perform a mass screening of contigs and identify the presence of antibiotic resistance genes. The user can choose which database to search from a list of available antimicrobial resistance databases.
The name ‘ABRicate’ was chosen as the first 3 letters are a common acronym for ‘Anti-Biotic Resistance.’ The word also has the form of an English verb, which suggests the tool is actually taking action against the problem of antibiotic resistance, and it is uniqueit unique enough to be easily searchable.
The tool is widely available to anyone who needs it, and can be found in many repositories. It is one of over 10,000 tools available in the Galaxy Toolshed. There is a tutorial available on the Galaxy Training Network which supports the use of this tool: Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition.
Torsten was recently awarded The Chan Zuckerberg Initiative’s Essential Open Source Software for Science grant for the development of another tool, Snippy. This open-source software tool was written to rapidly identify genetic differences between bacterial genome sequences, and work is underway to allow Snippy to support the latest genome sequencing technologies and viral and fungal pathogens too.
In recognition of the ongoing important contributions that Torsten makes, an award was established in his name. The annual ABABCS ‘Torsten Seemann Outstanding Bioinformatics Software Developer Award’ (previously sponsored by the ARDC) recognises an outstanding EMCR bioinformatic software developer from the Australian community with a view to promoting further efforts to develop and share bioinformatics methodologies.
Are your essential tools available on Galaxy Australia? Request an installation by the Galaxy Australia team if there’s a tool you think should be made available for everyone to use.
Nominate your favourite tool’s author for the Torsten Seemann Outstanding Bioinformatics Software Developer Award.
Boosting genomic medicine in Sri Lanka
A workshop in Sri Lanka taught medical doctors, research lab workers and university students to use Galaxy Australia for genomic medicine.
A group of Sri Lankan medical doctors, research lab workers and university students have just completed a hands-on bioinformatics training program using Galaxy to learn about genomics. Galaxy Australia team member, Dr Nuwan Goonasekera, joined an international panel of experts to deliver the workshop Bioinformatics for Genomic Medicine.
The US Embassy in Sri Lanka and Genelabs Medical offered the three week course to empower Sri Lankan genomics researchers to independently process and analyse DNA and RNA sequencing data. Participants gained the skills needed to identify clinically actionable mutations, helping to guide treatment plans based on patient data.
Dr Shurjo Sen from the NIH National Human Genome Research Institute (NHGRI) Office of Genomics Data Science requested Nuwan teach the 28 participants to use Galaxy in collaboration with the University of Colombo. The Galaxy Australia service underpinned the training while trainees were learning how to use Galaxy for sequence analysis and Nuwan made good use of Galaxy Australia’s Training Infrastructure as a Service (TIaaS). They also accessed the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL) to learn cloud computing and variant interpretation techniques. Use of this cloud-based genomic data sharing and analysis platform, along with Galaxy, will empower these Sri Lankan genomics researchers to access and analyse large genomic datasets in a collective and collaborative way.
Researchers come together to tackle the bioinformatics of fungal genomics
A diverse group representing commercial enterprise, academia, government research and citizen scientists gathered at ANU for a hands-on fungi bioinformatics workshop. The skills uplift was designed to support Bioplatforms Australia’s Functional Fungi and Plant Pathogen Omics National Initiatives.
Bioplatforms Australia brought together members of the Functional Fungi and Plant Pathogen Omics National Initiatives for a hands-on fungi bioinformatics workshop. Recognising that high throughput sequencing is the new normal for fungi research, hosts Prof Benjamin Schwessinger (Australian National University) and Dr Alistair McTaggart (Psymbiotika Lab) guided 38 researchers through theory sessions, hands-on practice, and real examples of applying bioinformatics to fungal research. Attendees were of diverse backgrounds in commercial enterprise, academia, government research and citizen science.
Each theory session was followed by a hands-on demonstration using Galaxy Australia. For example, Dr Mareike Möller (ANU) led a theory session on fungal genome assembly covering key concepts such as why long reads are essential for assembling complete genomes, and why genome assembly is like tackling the world’s hardest jigsaw puzzle. This was followed by Dr Anna Syme’s demonstration of genome assembly on Galaxy Australia. Anna wrote six dedicated workflows for fungi, which are now publicly available for any Australian to reuse or adapt on Galaxy Australia. To access them, head to Public Workflows and filter by tag ‘fungi’. If you’re just getting started with workflows, watch Anna’s Top Tips for using Galaxy workflows.
Benjamin and Alistair ensured the workshop was directly relevant to real-world fungal and plant pathogen research. All demonstrations used fungi data shared by Alistair, plus Dr Tara Garrad and Dr Kelly Hill from the South Australian Research and Development Institute, which remains available in the Bioplatforms Australia Data Portal.
Workshop participants felt that the mix of hands-on and theory sessions was ideal; demystifying the technology and bringing clarity to the broad array of options available in Galaxy Australia. Workshop materials and metadata are available in the Australian BioCommons Training Materials Zenodo repository.
Australian Outpost of BioHackathon Europe connects researchers to help shape global bioinformatics advances
The latest edition of the Australian Outpost of ELIXIR’s BioHackathon Europe brought together a group of researchers in Brisbane to help shape global bioinformatics advancements while networking with other life scientists, bioinformaticians and developers both at home and abroad.
A diverse collection of Australian researchers gathered in Brisbane for the latest edition of ELIXIR’s BioHackathon Europe. Thanks to the ongoing collaboration between Australian BioComons and ELIXIR, Australian life scientists had a powerful opportunity to shape global bioinformatics advancements, while networking with other life scientists both at home and abroad.
From 4 - 8 Nov, 18 keen Aussies worked from 12 - 9:30 pm each day to allow overlap with the European participants. This year our group worked across three projects:
One team developed a prototype data model converter that makes it easy to switch between data models for a given biomedical cohort. The team built mappings between common data formats OMOP and FHIR and central ‘meta cohort’ format, plus made a front end interface that enables data upload, selection of the data model, selection of the data format, and download of the converted dataset. This team was highly collaborative, with members from the BioCommons Human Genome Informatics team, software and bioinformatics engineers from Children’s Cancer Institute (CCI), bioinformaticians and data scientists from the Sydney Informatics Hub, and a University of Melbourne Masters of Genomics and Health student.
A particularly diverse team with representatives from BioCommons, Melbourne Bioinformatics, Curtin University, University of Sydney (Threatened Species Initiative), University of Tasmania, Symbio Labs, and QUT contributed to the FAIRtracks metadata schema. Their wide range of experience across conservation, agriculture and biodiversity was a new angle for a project that had previously focused on human data. The Aussies built many new connections and had a strong influence on the extension of the FAIRtracks schema into the life sciences.
Another team worked on was a key component of a larger European effort to ensure the global Galaxy project is sustainable and resources remain clearly accessible. Building the ‘Galaxy Codex’ led to major improvements in the findability of Galaxy resources such as tools, workflows and training material across microbiology, single cell omics, and proteomics.
Outside of the project work, the local team spent plenty of time getting to know each other and building their professional relationships, plus ate plenty of Spanish food to feel more connected to the Barcelona event! Dr Tim White, Data Science Software Engineer at the Sydney Informatics Hub, found collaborating in real time invaluable:
Typically in software development we are presented with the final product and don’t get to see the drafting process. It was fantastic to collaborate with the CCI software engineers to develop the code together and share what we consider at each stage of development.
Meanwhile, Dr Christian Fares, Senior Software Engineer at CCI, said their team found significant benefit from working outside of their usual area:
Our day to day roles wouldn’t normally encompass data model conversion, so we gained fantastic experience in a new area. We’ve been intending to learn more about the FHIR data standard, so attending the Aussie Outpost was a great opportunity, and we’ll be sharing our learnings with the rest of the CCI team soon.
We'll be bringing together another team for the next BioHackathon Europe in Nov 2025 - keep your eyes peeled for our call for applications mid next year! In the meantime, read ELIXIR’s summary of BioHackathon Europe 2024.