News
Subscribe to the Australian BioCommons monthly newsletter or read previous editions
Taking Nextflow to the next level: HPC Workshop applications now open
A new workshop will build practical skills to configure, optimise, and troubleshoot Nextflow pipelines for efficient and scalable execution on High Performance Computing (HPC) systems. It builds on the success of a recent national in-person training event attended by 90 life science researchers.
Life sciences research increasingly depends on analysing large and complex datasets. Tools like the workflow management system, Nextflow, can make this process reproducible, scalable, and efficient. But for many researchers, getting started with Nextflow can feel like a daunting leap.
Earlier this year the Nextflow for Life Sciences workshop introduced the fundamental principles of Nextflow pipeline development and guided newcomers to build reproducible and scalable scientific workflows with Nextflow, creating a multi-sample Nextflow workflow for RNAseq data preparation as an example. Led by trainers from the Australian BioCommons team at SIH, University of Sydney and supported by the National Bioinformatics Training Cooperative, 90 learners gathered at satellite sites in Sydney, Melbourne, Brisbane, Adelaide, Canberra and Perth, giving researchers the chance to connect locally with peers while benefitting from the national-level training.
Participants praised the balance of theory and practice, and nearly 90% of attendees rated the content as “very good” or “excellent”. A key highlight was the use of research-relevant datasets that empower scientists to apply their new skills directly to their work. As one participant reflected, “I got lots of knowledge on how to build my own Nextflow pipeline. The RNAseq example was fantastic - so many workshops use ‘Hello World’ examples that are impossible to scale up.”
We are excited to announce that applications are now open for our follow up workshop Nextflow on HPC to be held in November. This hands-on workshop will will provide participants with the practical skills to configure, optimise, and troubleshoot Nextflow pipelines for efficient and scalable execution on High-Performance Computing (HPC) systems.
Participants can expect to learn how to identify the differences between traditional HPC job submission and workflow execution via Nextflow, and how to best configure and execute scalable Nextflow workflows on HPC systems. Managing software environments using tools like singularity, and adapt these to fit within different HPC ecosystem constraints will be covered, along with tips on troubleshooting Nextflow workflows on HPC systems.
To make the most of this workshop you’ll need to have experience running simple Nextflow pipelines, and be looking to scale up your workflows to an HPC environment. You will be supported with access to national Tier 1 computing resources at Pawsey and NCI, so you must be associated with an Australian organisation.
These Nextflow workshops are part of Australian BioCommons’ mission to make advanced bioinformatics tools more accessible to researchers across the country. By combining national expertise with local support, this training is helping build capacity and strengthen the life sciences community.
Find out more about the workshop and submit your application: Nextflow on HPC.
Applications close 3 November 2025.
Thousands join international Galaxy training events
The Galaxy platform is celebrating its 20th anniversary this year and Galaxy Australia is one of the core BioCommons services. We are proud to provide and support a variety of opportunities for researchers to engage with the team, community and tools.
The Galaxy platform is celebrating its 20th anniversary this year. This collaborative data analysis platform is widely used by scientists around the world, and underpins many computational biology services. Galaxy Australia is one of the core services BioCommons delivers, so naturally we provide lots of opportunities for researchers to engage with the team, community and tools.
The Galaxy Australia team recently supported the Singapore Biology League, who chose to incorporate Galaxy into their online collaborative biology contest for the first time. This massive event welcomed over 2000 pre-university students who formed teams to run 2054 tools on Galaxy Australia over 4 hours. It was a great opportunity to broaden participants’ exposure to biology and bioinformatics beyond the school curriculum.
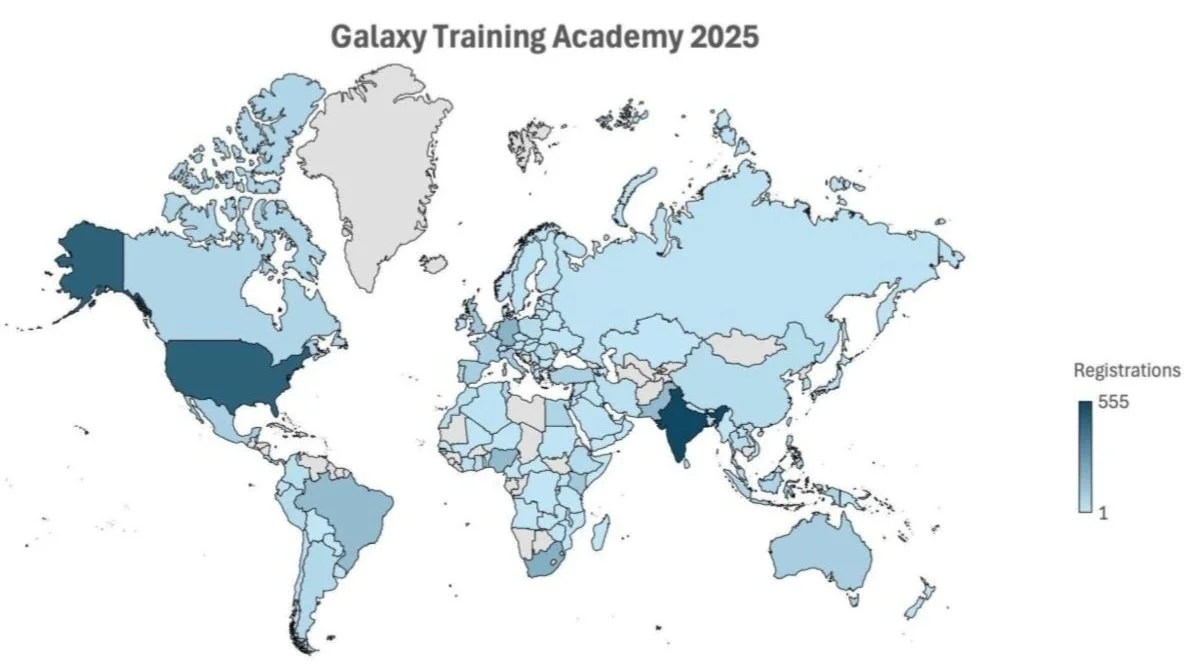
The week-long Galaxy Training Academy attracted more than 3500 people from across the globe this year. Seventy three Australian researchers were amongst the international cohort joining from their homes and offices to work through tutorials, with support as they needed it. Live help was provided in our time zones by Galaxy Australia’s Dr Anna Syme, Dr Igor Makunin and Dr Tristan Reynolds, who fielded questions on a wide range of topics from fungal genomics to bat ecology! Participants brought their own specialities as they learned how to use the fully subsidised Galaxy Australia platform for proteomics, genome assembly, transcriptomics, single cell RNAseq, microbiome analysis, machine learning and more.
In the lead up to the Galaxy Training Academy, BioCommons’ Dr Tiffanie Nelson and Galaxy Australia’s Dr Tristan Reynolds helped researchers understand what’s possible by presenting the webinar: No code, no problem: data analysis for biologists with Galaxy Australia. Examples included how Galaxy Australia is being used for biosecurity screening, foodborne pathogen detection and building reference genomes of the critically endangered swift parrot, as well as a tour of the practical features of Galaxy Australia that make sophisticated workflows like these accessible to all, regardless of their computational skills.
Get started by exploring the tutorials available: Galaxy Training Network
If you’re an existing Galaxy Australia user, improve your methods with our Top Tips videos
Stop the rot! Plant Bacteriologist’s genome assembly becomes Galaxy Australia’s 11 millionth job
An enthusiastic new user recently submitted the lucky 11 millionth data analysis job to the Galaxy Australia platform. Plant Bacteriologist Dr Toni Chapman has begun regularly using the fully-subsidised service for her genome assemblies of bacteria important to agricultural plant biosecurity and production.
Image courtesy of Dr Toni Chapman
An enthusiastic new user recently submitted the lucky 11 millionth data analysis job to the Galaxy Australia platform. Plant Bacteriologist Dr Toni Chapman has begun regularly using the service for her genome assemblies of bacteria important to agricultural plant biosecurity and production.
As a Senior Research Scientist in Agriculture and Biosecurity at the NSW Department of Primary Industries and Regional Development (DPIRD), Toni’s work spans both diagnostics and research. Using NovaSeq and HiFi sequencing technologies, she assembles bacterial genomes to help growers diagnose the cause of disease symptoms in their plants or to identify bacteria present in suspect samples collected by biosecurity officers.
In the past, Toni needed to rely heavily on bioinformaticians for assistance with genome assembly of the bacteria she works with. Now after attending a training workshop and signing up for access to the fully-subsidised Galaxy Australia service, she independently completes more of the analysis steps and is able to identify the bacterial pathogens herself.
“Over time I have been learning to code and to use various programs for genome assembly, but only returning to it on a casual basis makes it very hard to maintain the necessary skills in the bioinformatics space. Being able to design workflows in Galaxy Australia that I can come back to at any time makes assembling genomes easy, even if there has been weeks or months between visits.”
Toni contributes to the Plant Pathogen ‘Omics Initiative, which is generating molecular reference data for plant pathogens in Australia. Established by Bioplatforms Australia to support research in plant protection and enhance biosecurity surveillance efforts, the national plant pathogen community is collaborating to create high quality data that can be shared with all researchers via the Data Portal. Toni was one of a large group that came together with members of the Functional Fungi and Plant Pathogen ‘Omics National Initiatives for a hands-on bioinformatics workshop, learning how to use Galaxy and the programs needed for genome assembly using their own real-world scenarios.
As part of her contribution to the Initiative, Toni is conducting genome assembly on Pseudomonas species that cause disease in plants, to update the taxonomy of collection isolates and gain a more accurate view of which pathogens are in Australia. In another project, she is sequencing the bacterial pathogens that cause soft rot disease in plants. The incidence of soft rot is increasing, and so is the range of bacteria that can cause the disease. The newly assembled genomes of these bacteria are updating existing culture collection identities and helping to understand the diversity of bacteria that cause soft rot infections in Australia.
If you’d like to find out more about her work, see Dr Toni Chapman’s publications.
If you are interested in hearing about the different types of research that Galaxy Australia gets used for, watch this recorded webinar: No code, no problem - data analysis for biologists with Galaxy Australia.
Join a Metagenome Assembled Genomes hackathon
The “Optimising MAGs-building workflows hackathon” is taking place in October and we want to know if you’d like to join! An international group is participating in Europe, and BioCommons is planning to offer a complementary event in Australia. Please join us if you are interested in enhancing MAGs-building workflows, developing user-friendly training materials, advancing workflow evaluation methods, or building intelligent computational resource estimation tools.
The Optimising Metagenome Assembled Genomes building workflows hackathon is taking place in October and we want to know if you’d like to join!
Multiple ELIXIR nodes and the international Galaxy community are coming together in Europe to participate, and BioCommons is planning to offer a complementary event in Australia.
What is this?
Hackathons involve collaborative group work with people outside your normal network, solving problems around a shared topic of interest within a limited period of time. The aims of the event are:
Enhancing FAIR MAGs building Workflows
Developing user-friendly training materials
Advancing workflow evaluation methods (using CAMI infrastructure & real data)
Building intelligent computational resource estimation tools
The event takes place 6-9 Oct 2025. It will run in person in Freiburg, Germany and online. If there is enough interest, BioCommons will organise an in-person event in Australia to overlap with the European event.
What’s in it for me?
Collaborative group work: You'll work with people outside your usual network.
Expert connections: You'll connect with a national and international group of experts and enthusiasts.
Workflow enhancement: You'll contribute to enhancing MAG workflows.
Training material development: You'll participate in developing related training materials.
Skill development: While specific skills are not required, you can enhance your knowledge of microbiome data analysis or MAGs.
International participation: Opportunity to participate in an international hackathon with colleagues in Europe.
Who can join in?
It is open to anyone who wants to build MAGs or anyone with a general interest in MAGs, microbiome analysis and the associated training to run MAG workflows. To join in the hackathon, some knowledge of microbiome data analysis or MAGs is preferred. There is no cost to join.
How would this work?
Australian BioCommons regularly run satellite events that facilitate participation in global hackathons. If there is interest, we would ideally work together in person in Australia. Given European business hours coincide with Australian evenings, we would connect with the European hackathon team during some overlapping hours in the afternoon and work asynchronously for the other hours.
Are you interested in joining in?
If you think that you might be interested to join in this hackathon event, please get in touch with Tiff Nelson by 20th June 2025.
Nextflow workshop combines benefits of hands-on training and community building
BioCommons’ Nextflow for the life sciences workshop heralds a return of our dispersed model of hands-on training. By connecting supported in person satellite sites with online trainers this workshop enables access to Nextflow experts and fosters local connections that are essential for continued learning.
BioCommons’ Nextflow for the life sciences workshop heralds a return of our dispersed model of training that combines the benefits of in person and online events to enable access to experts and foster local connections that are essential for continued learning. First pioneered in 2019, this model has been successful in ensuring scalable and more equitable delivery of short-course bioinformatics training across Australia and has been adapted internationally.
Nextflow for life sciences workshop participants will join in person satellite sites at host universities and research institutes across Australia where they will connect with peers and be supported by experienced local facilitators as they put their new Nextflow skills into action. Each of these sites will connect online with Nextflow experts and lead trainers Fred Jaya and Dr Michael Geaghan at the University of Sydney’s, Sydney Informatics Hub who will introduce key concepts and demonstrate how to use fundamental Nextflow elements to develop, execute, and debug a scalable multi-step life science workflow.
Find out more and apply for the workshop. Applications close 27 June 2025.
This workshop is made possible by an exceptional network of facilitators and trainers from the national Bioinformatics Training Cooperative.
New seminar series ignites curiosity and builds practical AI skills in the life sciences
A series of webinars will feature Australian researchers from across academia and industry sharing their experiences of using AI in the life sciences. Hear how AI is being used to push boundaries, solve problems, and reimagine the way we do science. From multi-omics analysis to drug discovery, structural biology and the ethics of AI in science, these free events will explore diverse and practical applications across the life sciences, focusing on the stories, insights, and experiences behind the research.
AI is reshaping life sciences by enabling researchers to analyse complex datasets, automate workflows, and gain deeper insights into biological processes. Australian BioCommons is supporting the community to adopt these technologies through a series of training events that explore what’s possible and build skills in using AI effectively.
We’ve invited a diverse range of your peers from across academia and industry to share their experiences of using AI in the life sciences. Between June and September you’ll hear how AI is being used to push boundaries, solve problems, and reimagine the way we do science. From multi-omics analysis to drug discovery, structural biology and the ethics of AI in science, they will explore diverse applications across the life sciences, focusing on the stories, insights, and experiences behind the research.
In August, our online workshop Machine learning in the life sciences will provide a hands-on opportunity to compare and contrast commonly used algorithms for constructing predictive models, explore some of their trade-offs and identify types of scenarios in which they can be applied.
In case you missed it, Australian BioCommons’ AI Technical Lead, Dr Benjamin Goudey, recently broke down AI concepts, clarified key terminology, and showcased real-world examples in this recorded webinar: Deciphering AI for the Life Sciences.
AI is a fast evolving field so rest assured that further events are in the pipeline.
Building bridges for better data sharing: ENA experts empower Australian researchers in data submission
A visit from the European Nucleotide Archive (ENA) team enhanced Australian researchers’ skills in submitting and retrieving genomic, metagenomic, and environmental DNA (eDNA) data to/from international repositories. The two weeks together provided a unique opportunity to engage directly in our time zone through an intensive series of workshops and roundtable discussions.
Participants of the in-person roundtable came from around Australia to meet the ENA team
For two weeks in March and April 2025, Australia’s life sciences community had a unique opportunity to engage directly with the European Nucleotide Archive (ENA) team. In a first-of-its-kind initiative, Dr Joana Pauperio (Biodiversity Curator, European Nucleotide Archive, EMBL’s European Bioinformatics Institute) and Maira Ihsan (User Support Bioinformatician, European Nucleotide Archive, EMBL’s European Bioinformatics Institute) visited Australia to deliver an intensive series of seven workshops and four roundtable discussions, aiming to enhance Australian researchers’ skills in submitting and retrieving genomic, metagenomic, and environmental DNA (eDNA) data to/from international repositories.
Organised by Australian BioCommons, the visit built technical capacity and opened a direct dialogue between the ENA and the Australian research community about the future of data submission, retrieval, and brokering. High-quality data submission to international archives like the ENA ensures that Australian-generated genomic and environmental data can contribute to global research efforts. Yet, challenges in submission processes, metadata preparation, and understanding of repository workflows can act as barriers. Bringing ENA experts together in person allowed Australian researchers to receive tailored, hands-on guidance, overcoming time zone challenges and helping the ENA team witness firsthand the hurdles local researchers face.
Workshops: Hands-on learning and capacity building
Across six data submission workshops, participants learned various data submission pathways (e.g., via Webin-CLI, programmatic, and command line) to submit:
Raw reads, genome assemblies, and annotations
Metagenome-Assembled Genomes (MAGs)
Environmental DNA (eDNA) data
A data retrieval workshop provided an opportunity for participants to practice retrieving different data types from the ENA using various tools and protocols.
Feedback was welcome at all times by providing a living document for queries that were addressed during and after the workshop, and breakout rooms for 1:1 discussions were available.
Roundtables: Listening to the community
One in-person and three online roundtable discussions were also hosted to facilitate direct communication between ENA and Australian researchers.
In-person Roundtable
This meeting between invited members of Bioplatforms Australia, Bioplatforms Australia Data Portal, Australian Reference Genome Atlas (ARGA), the Australian Tree of Life project, and the ENA teams focused on information exchange and potential collaboration in the global biodata landscape. Key topics included data brokering to ENA, species taxonomy, and the possibility of establishing an Australian node within the International Nucleotide Sequence Database Collaboration (INSDC). The immediate next step identified was to further explore data brokering. The roundtable provided a valuable forum for discussing opportunities and challenges in collaborating with the ENA and enhancing Australia's contribution to international data repositories.
Genomics Roundtable
The meeting facilitated discussions on topics including Genome assembly and annotation efforts at scale in Australia, ENA's role as a global repository and challenges in annotation submissions to INSDC. It aimed to improve understanding of data publication options and ENA submission processes.
MAGs Roundtable
The meeting facilitated discussions on topics including the use of MAGs in Australia, the role of ENA+MGnify as a global repository, challenges in mass submission of MAGs, issues with submitting MAG data for organisms not represented in the NCBI Taxonomy, and suggestions for improvement.
eDNA Roundtable
The meeting facilitated discussions on topics including eDNA use across various sectors, Australian eDNA reference library initiatives like the National Biodiversity DNA Library (NBDL), making eDNA data FAIR (Findable, Accessible, Interoperable and Reusable) and the ENA as a global repository for eDNA data, data interoperability between resources, and data sharing with third-party platforms like GBIF.
Looking ahead
The momentum generated by the workshops and roundtables will continue through:
The creation of self-paced training materials: by converting the workshop content and hosting it on the EMBL-EBI training website to ensure researchers have access to training when they need it
Efforts to explore an Australian data brokering pathway as part of the Australian Tree of Life (AToL) project
Strengthened connections between Australian researchers and INSDC repositories
By bridging expertise across continents, the collaboration between ENA and the Australian life sciences community is helping ensure that Australian research continues to have a strong, visible impact on the global stage.
Join 41,000 users of this fully-subsidised and easy to use Australian bioinformatics platform
Galaxy has proven to be such a versatile data analysis platform that Galaxy Australia supports over 41,000 users. Now is the perfect time to get on board with Galaxy, with a variety of ways to get involved on offer in the coming months.
Galaxy has proven to be such a versatile data analysis platform that Galaxy Australia now supports over 41,000 users. This fully subsidised, open-source system for analysing and visualising data, authoring workflows, training, publishing tools, and managing infrastructure is powered by a world-wide community of 500,000 Galaxy users as well as the institutions who invest in its growth. Now is the perfect time to get on board with Galaxy, with a variety of ways to get involved on offer in the coming months.
If you are curious to see how Galaxy is being used in a wide range of research settings, come along to our upcoming webinar showcasing how Galaxy Australia is supporting genome assembly and annotation, metagenomics, proteomics, transcriptomics, data visualisation and more. Register now for No code, no problem - data analysis for biologists with Galaxy Australia on Tue 29 Apr 2025.
The global community’s annual training event - Galaxy Training Academy - is coming up in May. This week-long, completely free, online training event will help you master the Galaxy platform for data analysis. It is self-paced and you can choose from topics including Proteomics, Assembly, Transcriptomics, Single Cell, Microbiome or Machine Learning data analysis in Galaxy. The Galaxy Australia team will be on hand to answer any questions you have during the event over 12-16 May.
Most people simply make use of the platform to do their research, but there’s an open invitation to contribute to this lively community too. The international Galaxy Community,has put together a Get Started page to help you find out more about terminology, resources, servers, types of communities, and the different ways to contribute to the great things the Galaxy community achieves together. You can also connect with the community in person at the Galaxy and Bioconductor Community Conference 2025 is coming up in June in the USA.
Strategic partnerships formed around the platform are key in making it readily available to Australian researchers. Australian BioCommons is able to offer the service at no cost to researchers because of the commitment of valuable partners QCIF, The University of Melbourne and AARNet who help manage the Galaxy Australia service and provision of computational resources provided by respected national institutions and providers AARNet, ARDC Nectar Research Cloud, the University of Melbourne, QCIF, National Computational Infrastructure, Pawsey Supercomputing Research Centre and Microsoft Azure. These efforts are backed by funding from the University of Melbourne, the Queensland state government and Bioplatforms Australia, who bring Federal investment through the National Collaborative Research Infrastructure Strategy.
If you’re ready to start using Galaxy Australia, you can jump right in and try it now: usegalaxy.org.au/
Addressing DNA sequencing data submission challenges at global scale: your invitation to learn from the experts
A range of special events for Australian researchers will focus on streamlining the data submission processes for various data types, addressing common challenges, and providing best-practice guidance on interacting with the European Nucleotide Archive (ENA).
Data submission to international repositories is an essential task for researchers, but it can often come with challenges. A range of special events for Australian researchers will focus on streamlining the data submission processes for various data types, addressing common challenges, and providing best-practice guidance on interacting with the European Nucleotide Archive (ENA).
BioCommons is partnering with specialists from the ENA and the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) to deliver a series of hands-on workshops and online community discussions to help Australian researchers share their valuable data. The ENA is committed to maximising the impact of the ENA platform, sequencing technology and data, and will travel to Australia to support local researchers' data submission and discuss our particular needs.
The two-week program in late March and early April will feature seven targeted workshops covering submission and retrieval workflows for specific data types (raw reads, assembled and annotated genomes, metagenome assembled genomes (MAGs) and environmental DNA (eDNA)). Three interactive online community discussions around the same themes round out the program and offer the opportunity for Australian researchers to share their experiences, challenges and potential solutions directly with international specialists from the ENA.
ENA is a globally recognised open platform for the management, sharing, integration, archiving and dissemination of nucleotide sequence data. This comprehensive resource preserves the world’s public-domain output of sequence data and is an essential tool for many researchers, but sometimes people can struggle with using it. Given that submission of omics data and associated contextual metadata to global data repositories is considered best practice and is mandated by science publishers, we want to enable researchers to facilitate the long-term preservation, findability and reusability of their data.
This is a unique opportunity to gain invaluable insights and practical skills from leading experts in omics data management and submission. Whether you're a seasoned researcher or just starting out, these workshops and discussions will empower you to confidently navigate the complexities of data sharing.
'Roundtable' Community Discussions: Challenges and solutions
01 - 03 Apr, 10:00-12:00 AEDT
Join our online 'Roundtable' Community Discussions with members of the ENA team.
Each session will focus on a different theme: Genomics - genome annotations and assemblies, and pangenomes; MAGs - submitting MAGs at scale, and how to submit MAG data when there are gaps in the NCBI species taxonomy; and eDNA - recent changes enabling eDNA sequences to be more findable through submission to ENA. Register here for roundtables.
Hands-on Training workshops: Data submission and retrieval
25 Mar - 03 Apr, 13:00-16:00 AEDT
Learn directly from the ENA team in a range of practical online training workshops to explain genome assembly and annotation, MAG and eDNA data submission and retrieval.
Our community consultations teased out some of the challenges faced by Australian-based researchers in the data submission process, and BioCommons published a set of recommendations to address them. One ambition of a range of ongoing activities around interfacing with international omics data repositories is to facilitate improvements by connecting the right people.
The European Nucleotide Archive (ENA) is a Global Core Biodata Resource, an ELIXIR Core Data Resource and is hosted by EMBL-EBI. It is one of the three nodes of the International Nucleotide Sequence Database Collaboration (INSDC).
INSDC is actively seeking additional members to enhance Global Participation.
Boosting genomic medicine in Sri Lanka
A workshop in Sri Lanka taught medical doctors, research lab workers and university students to use Galaxy Australia for genomic medicine.
A group of Sri Lankan medical doctors, research lab workers and university students have just completed a hands-on bioinformatics training program using Galaxy to learn about genomics. Galaxy Australia team member, Dr Nuwan Goonasekera, joined an international panel of experts to deliver the workshop Bioinformatics for Genomic Medicine.
The US Embassy in Sri Lanka and Genelabs Medical offered the three week course to empower Sri Lankan genomics researchers to independently process and analyse DNA and RNA sequencing data. Participants gained the skills needed to identify clinically actionable mutations, helping to guide treatment plans based on patient data.
Dr Shurjo Sen from the NIH National Human Genome Research Institute (NHGRI) Office of Genomics Data Science requested Nuwan teach the 28 participants to use Galaxy in collaboration with the University of Colombo. The Galaxy Australia service underpinned the training while trainees were learning how to use Galaxy for sequence analysis and Nuwan made good use of Galaxy Australia’s Training Infrastructure as a Service (TIaaS). They also accessed the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space (AnVIL) to learn cloud computing and variant interpretation techniques. Use of this cloud-based genomic data sharing and analysis platform, along with Galaxy, will empower these Sri Lankan genomics researchers to access and analyse large genomic datasets in a collective and collaborative way.